A Novel CRISPR-Cas-Triggered Isothermal Amplification Method for Ultrasensitive DNA Detection
Date:12-12-2018 | 【Print】 【close】
Polymerase chain reactions (PCR) is a technique generally used in molecular biology to make copies of a specific DNA segment, but due to its limitation of power-hungry thermocycling and operational complexity, an efficient isothermal DNA amplification tools are of great importance to basic research as well as diagnostic/monitoring applications.
Although several isothermal methods have been proposed, they are suffered from trade-offs with sensitivity, specificity, simplicity and cost. Moreover, some of these techniques still require an initial heat-denaturation step to unwind the double-stranded DNA (dsDNA) targets thereby further limiting their applications.
In this respect, a true isothermal DNA amplification/detection method with attomolar sensitivity, single-base specificity and simple reaction scheme is highly desirable.
A research team led by Dr. ZHOU Wenhua and supervised by Prof. YU Xuefeng at the Shenzhen Institutes of Advanced Technology (SIAT) of the Chinese Academy of Sciences has reported a novel isothermal approach for amplifying and detecting dsDNA based on a CRISPR/Cas9 (clustered regularly interspaced short palindromic repeat, and CRISPR-associated protein 9)-triggered nicking endonuclease-mediated Strand Displacement Amplification method (namely CRISDA) with attomolar sensitivity and single nucleotide specificity.
The CRISPR/Cas9 system originally derived from the adaptive prokaryotic immune system, now has widely been applied as a prominent tool in many fields. However, therapeutic potentials of CRISPR genome editing techniques are limited to cell- or animal-based models, due to the complexity of human organism and possible ethical controversies.
On the other hand, on account of the simplicity and high sensitivity/specificity, various CRISPR effectors including Cas9, Cas12a, and Cas13a/b have been explored and the prospect of CRISPR-based nucleic acids diagnostics (CRISPR-Dx) is encouraging.
All the CRISRP-Dx approaches reported so far require an initial amplification step such as PCR, NASBA and RPA to specifically amplify targeted nucleic acids and CRISPR effectors are only used in endpoint analyses. In other words, the performance of these CRISRP-Dx approaches is mainly determined by the initial amplification, while the superior sensitivity and stringent PAM recognition of CRISPR systems have not been fully exploited.
Thus, by taking full advantage of the high sensitivity, stringent specificity and unique conformational rearrangements of CRISPR effectors in recognizing the targeted DNA, the team developed the CRISDA technique for sensitive amplification and detection of double-stranded DNA (dsDNA). The established CRISDA technique exhibited attomolar sensitivity and single nucleotide specificity in amplification and detection towards targeted DNA fragments as well as targeted regions in the genomes of human and genetically-modified soybean MON87705.
This work named “A CRISPR-Cas9-triggered strand displacement amplification method for ultrasensitive DNA detection” has been published in Nature Communications.
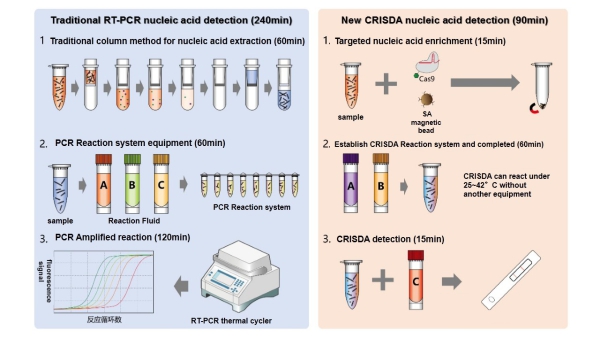
Figure 1. Comparisons between conventional PCR and CRISDA techniques
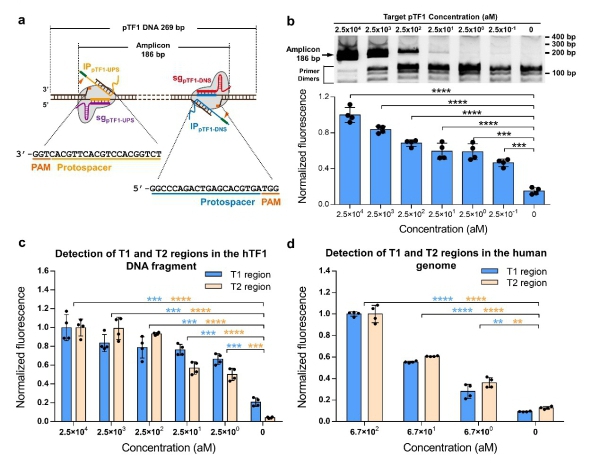
Figure 2. CRISDA-based DNA amplification and detection with attomolar sensitivity. (a) Schematic of CRISDA-based DNA amplification and detection towards a DNA target. (b) CRISDA-based detection towards a synthetic DNA fragment. (c) CRISDA-based detection towards a DNA fragment from the human genome. (d) CRISDA-based detection towards specific regions in the human genome
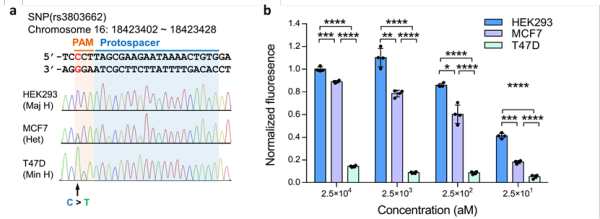
Figure 3. CRISDA-based DNA amplification and detection with single-nucleotide specificity. (a) Sequencing verification of a breast cancer-associated SNP. (b) CRISDA can discriminate SNP for highly sensitive human genotyping