Scientists Design A Synthetic Genetic Circuit to Quantify Repeat Deletion in Bacteria
Date:11-05-2020 | 【Print】 【close】
Repeat sequences (known as patterns of nucleic acids\DNA\RNA) are ubiquitous in the genome of prokaryotes and eukaryotes.
The rearrangement between direct repeats (or genetic sequence) can result in deletions or expansions of DNA sequences, which contributes the genetic plasticity, regulation of transcription and protein coding sequence variations.
Thus, quantification of a genetic rearrangement rate is a primary aspect in deciphering its underlying mechanisms.
The previous antibiotic resistance selection method, commonly achieved by inserting direct repeats within the coding sequence of tetR (resistance gene for tetracycline), has been used to quantify the direct repeat deletion (DRD) rate, but limited in false positive events, affected physiological process from the antibiotic, and the length of repeat arms (LRA).
Scientists from the Shenzhen Institutes of Advanced Technology of the Chinese Academy of Sciences (SIAT) and Huazhong University of Science and Technology (HUST) have designed a synthetic genetic circuit, termed TFDEC (which stands for Three color Fluorescence-based Deletion Event Counter), to quantify the DRD rate under neutral conditions. The study was published in ACS Synthetic Biology.
In this present study, the scientists designed a fluorescent logic gate-based method to quantify the DRD rate by distinguishing the deletion and mutation events from all detected events with fluorescent intensity.
They found that the DRD rate was RecA-dependent for long LRA (>500bp) and RecA-independent for short LRA (<500bp) in Pseudomonas aeruginosa. They also found that the increase of DRD rate followed an S-shaped curve with the increase of length of repeat arm, while treating cells with ciprofloxacin enhanced the DRD rate but did not change the LRA-dependence of DRD. In addition, the DRD rate increased for long LRA in the uvrD (8-fold) and radA (4-fold) mutants, which was related to RecA protein.
Scientists concluded that this study may be useful in the investigation of a persister emergence during antibiotic treatment and in unravelling the roles of certain proteins during stress-driven evolutions of bacterial population.
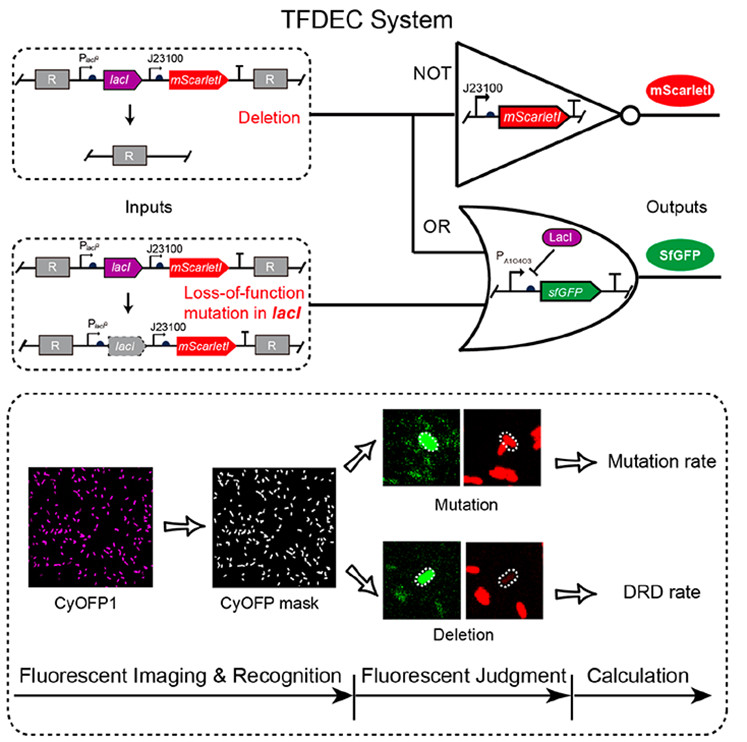
Schematic of gene circuit of TFDEC system. (Image by SIAT)
Media Contact: