SIAT Scientists Made Progresses on Saccharomyces cerevisiae's Biological Components Standardization and Applications in Metabolic Engineering
Date:16-06-2021 | 【Print】 【close】
Supported by the funding of the External Cooperation Program of Chinese Academy of Sciences, the research team lead by Dr. DAI Junbiao, Shenzhen Institutes of Advanced Technology (SIAT) of the Chinese Academy of Sciences, and the research team of Dr. CAI Yizhi, University of Edinburgh (current: University of Manchester), worked collaboratively in the standardization of yeast's biological components and its application in metabolic engineering. A series of advances have been achieved in recent years, including the development of 'Wicket' tools to achieve multi-copy integration of genes in a given metabolic pathway (Hou et al., 2018, ACS Synthetic Biology), design and construction of the artificial protein scaffold system (AProSS) to optimize metabolic flow (Li et al., 2018, Metabolic Engineering), and standardization of the assembly and quantitative phenotyping methods to improve the efficiency of constructing synthetic yeast chromosomes (Lin et al., 2019, ACS Synthetic Biology) etc. A total of 10 papers and 1 patent have been published, and the series of discoveries provided technologies and resources for researches on synthetic biology using Saccharomyces cerevisiae as the chassis.
Yeast can be used as a microbial cell factory to produce valuable chemicals. The introduction of exogenous metabolic pathways into specific positions of chromosomes in the form of multiple copies for stable expression is of great significance for increasing yield. With the funding of this project, the researchers used CRISPR/Cas9 technology to insert artificially designed "wicket" sequences into the yeast genome, making it possible to achieve integration of β-carotene synthesis pathway at an efficiency close to 100% with no requirement for any selective markers. The ability to integrate multiple copies of genes in a given pathway provides an effective tool for optimizing the yield of target products (Figure 1).
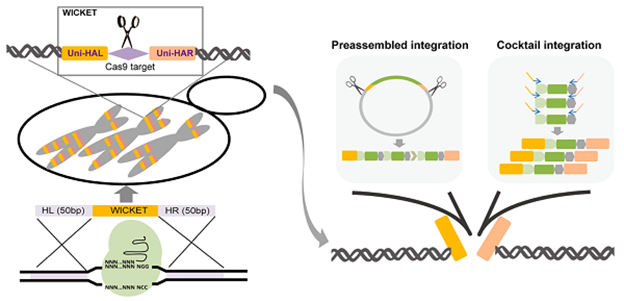
Figure 1. Integration of multiple copies of genes using Wicket
The quantitative relationship and spatial distance between the substrates in each reaction of the metabolic pathway have a significant impact on metabolic flow and final output of the target product. Funded by this project, the researchers established a new method (AProSS) to increase the yield of specific natural products in yeast by rapidly assembling artificial scaffold proteins. Through the modular design of protein domains and the changes in the type, number and location of the domains, the researchers successfully constructed scaffold proteins with different characteristics to regulate both the direction of metabolic flow and the yield of violacein and deoxyvioletin respectively by 29 % and 63% (Figure 2).
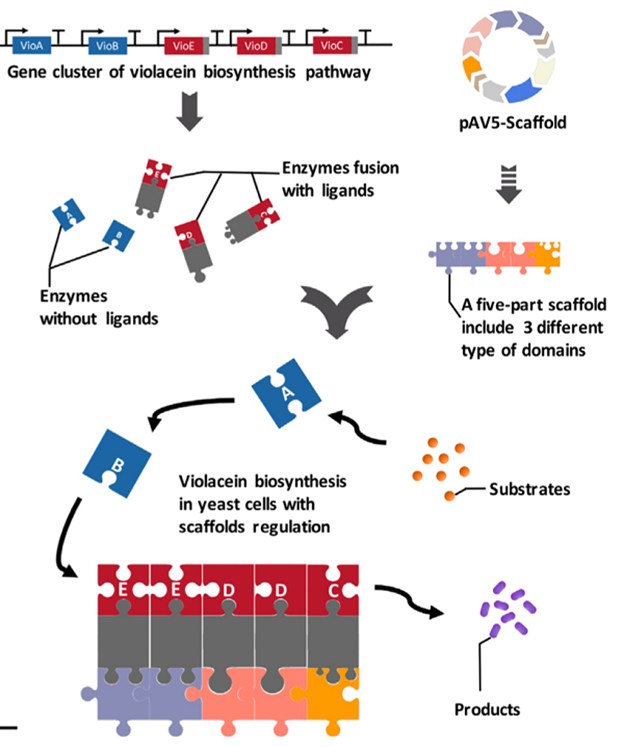
Figure 2. Using artificial scaffold protein to increase the production of violacein (Image by SIAT)
How to quickly construct and characterize artificial cells is an important issue in synthetic biology. In this project, the researchers developed a high-throughput, semi-quantitative phenotyping method to evaluate the growth of synthetic yeast by choosing the test conditions and standardization of statistical analysis methods. At the same time, the research team also designed an efficient chromosome assembly strategy based on CRISPR/Cas9-mediated Gene Conversion, which greatly saved the time for assembly of large-segment DNA. These two methods can improve efficiency in the process of characterization and construction of synthetic strains, which greatly promotes the development of synthetic genomics related projects, and is expected to provide an artificial chassis for yeast-based metabolic engineering (Figure 3).
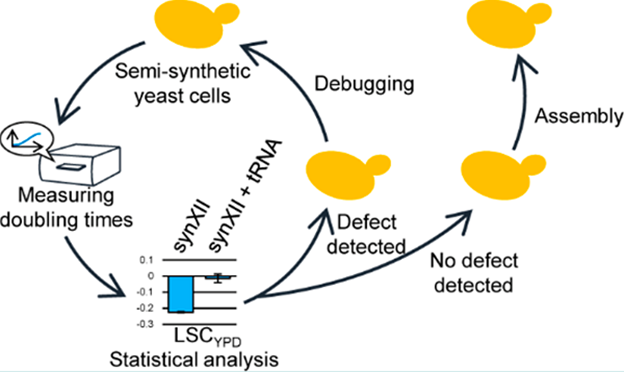
Figure 3. Using semi-quantitative phenotyping and fine assembly strategies to improve the efficiency of chromosome synthesis (Image by SIAT)
Media Contact:
SUN Lujia
Email: lj.sun @siat.ac.cn
Download the attachment: