[待上线] Accelerating Design of Engineered Living Materials through Integrated Technological Workflow
Date:04-07-2023 | 【Print】 【close】
The field of engineered living materials (ELMs) aims to develop biomaterials with "living" attributes like autonomous growth, self-healing, and environmental responsiveness, similar to natural living materials.
ELMs can be created in two ways: using engineered cells to incorporate novel functionalities (self-organizing living materials or biological ELMs), or by embedding living cells in an organic or inorganic matrix (hybrid living materials).
Self-organizing living materials mimic the properties of natural living materials and offer opportunities for advanced capabilities using engineered biological systems. However, their further development faces challenges due to the lack of engineerable chassis and programmable biopolymers in microorganisms, especially non-pathogens.
A research team from the Shenzhen Institute of Advanced Technology (SIAT) of the Chinese Academy of Sciences has developed an integrative technological workflow for ELMs by synergistically combining tools from bioinformatics, structural biology, and synthetic biology. The workflow enables the exploration, comprehension, and utilization of bacterial biopolymers of interest and suitable biopolymer-producing bacteria as structural building blocks, and then hosts for systematic and sequential design of living materials.
The results were published in Nature Chemical Biology in July **.
The team developed an integrative technological workflow for ELMs by rationally combining tools from bioinformatics, structural biology, and synthetic biology. Building on those tools, this technological workflow facilitates mining, understanding, and harnessing of bacterial biopolymers of interest and appropriate biopolymer-producing bacteria, as structural building blocks and hosts, for living materials design in a systematic and sequential manner.
The team embarked on the development of bioinformatics software, referred to as "Bacteria Biopolymer Sniffer" (BBSniffer), by harnessing various bioinformatics tools. BBSniffer utilizes an extensive library of bacterial genome sequences to assist in the search for biosynthetic gene clusters (BGC) of specific functional biopolymers, including proteins, polysaccharides, and other biopolymers. It subsequently generates a screened list of biopolymer-producing bacteria, comprising functionally useful and experimentally viable non-pathogens.
"Using an existing pathogenic pilus as input, we identified the biosynthetic gene cluster for covalently-linked pili (CLP) in the industrially significant bacterium Corynebacterium glutamicum," stated Prof. ZHONG Chao, the corresponding author of this study.
The team investigated CLP assembly in C. glutamicum, confirming that CLP fibers consist of minor pilins (Spa1 and Spa3) and a major pilin (Spa2) from the BBSniffer-mined CLP BGC. They found that Spa2 is essential for CLP assembly. Unlike Gram-negative bacteria, CLP monomer subunits are joined by intermolecular isopeptide bonds catalyzed by sortase, providing tensile strength. CLP subunits also contain auto-catalyzed intramolecular isopeptide bonds, enhancing robustness. Some pilin proteins in different CLP strains have additional disulfide bonds for further stability. Using liquid chromatography-tandem mass spectrometry (LC-MS/MS), they found intermolecular isopeptide and disulfide bonds in Spa2. These bonds play crucial roles in CLP formation.
The proteinaceous nature of CLP fibers allows genetic engineering for elaboration. The team identified fusion sites to append peptides/proteins to Spa2, discovering that sortase-mediated polymerization persists, enabling various proteins to be engineered into a programmable extracellular protein scaffold. Split components can also co-assemble in the extracellular CLP scaffold. Co-assembly of two fusion proteins in Spa2 suggests potential metabolic channeling applications. To degrade cellulose into glucose for lycopene production, they created a catalytic cascade of multiple cellulases in C. glutamicum.
This work demonstrates engineerable CLP biopolymer as a scaffold for anchoring multiple cellulases, leading to enhanced glucose yield and upcycling renewable waste into valuable chemicals by combining extracellular and intracellular bioconversion abilities.
Subsequently, CLP-enabled living materials facilitated the conversion of cellulosic biomass into lycopene by coupling extracellular and intracellular bioconversion abilities.
Subsequently, the engineering of CLP-enabled living materials facilitated the conversion of cellulosic biomass into lycopene by coupling extracellular and intracellular bioconversion abilities.
In this study, they established an integrative technological workflow combining bioinformatics, structural biology, and synthetic biology for efficient living materials design. The workflow allows rapid mining of biopolymer-producing non-pathogenic chassis, understanding the molecular assembly mechanism of biopolymers, and engineering biopolymer building blocks in a systematic manner. With this approach, one can search for biopolymer-producing strains in non-pathogenic industrial workhorses and design new ELMs from scratch, regardless of existing knowledge of endogenous biopolymers.
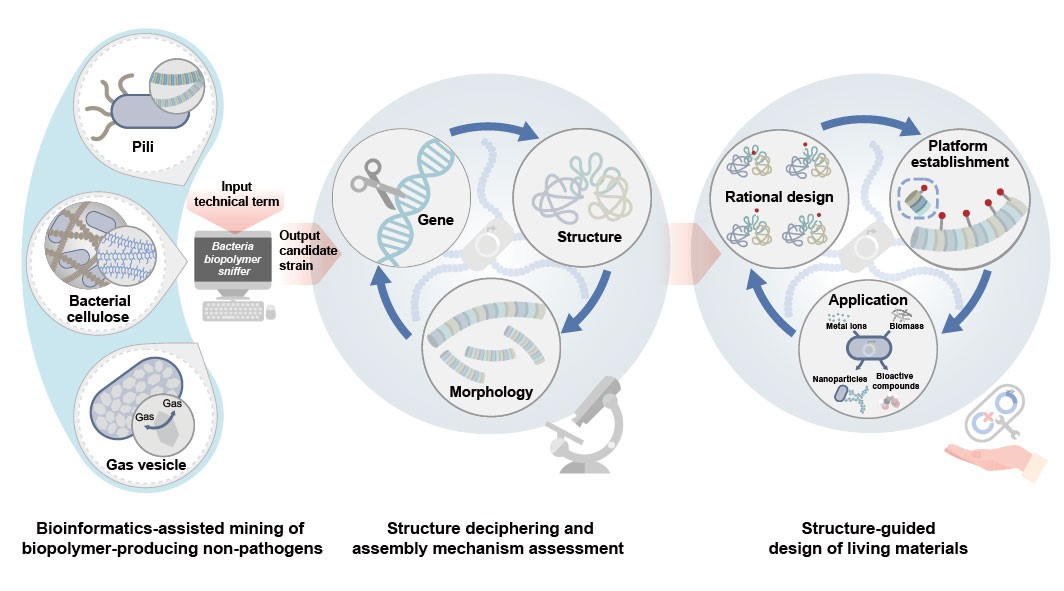
The integrated technological workflow combines tools from bioinformatics, structural biology, and synthetic biology to mine, comprehend, design, and engineer structural building blocks for living materials. (Image by SIAT)
Media Contact:
ZHANG Xiaomin
Email:xm.zhang@siat.ac.cn