Unlocking Cellular Mysteries: Advancements in Single-Cell Omics Analysis with Genome-Scale Metabolic Modeling
Date:05-03-2024 | 【Print】 【close】
The widespread applications of single-cell technologies have generated a large amount of single-cell data, resulting in a growing need for methods for single-cell data analysis and interpretation. While methods such as machine learning allow for single-cell data analysis, they cannot interpret the data very well.
Genome-scale metabolic models (GEMs) contain a priori metabolic network knowledge and have become a powerful tool for integrating and interpreting omics data from bulk samples. Therefore, GEMs may also be used to analyze single-cell data, and this field is indeed emerging. Integrative analysis of single-cell data with GEMs is expected to reveal new discoveries in cell metabolism and explore metabolic differences between cell types, even within complex tissues.
Recently, Dr. CHEN Yu from the Shenzhen Institute of Advanced Technology (SIAT) of the Chinese Academy of Sciences and his collaborator Dr. Eduard J Kerkhoven from Chalmers University of Technology, Gothenburg, Sweden, retrospectively examined the methods developed for integrating bulk omics data with GEMs.
The result was published in the Current Opinion in Biotechnology entitled "Single-cell omics analysis with genome-scale metabolic modeling."
The integration of single-cell data into GEMs appears to be quite simple at first sight since it only needs to adapt methods that have been developed for bulk data to single-cell data. Therefore, this article first introduces two categories of common methods for integrating bulk omics data with GEMs. The first one is model extraction, which basically removes inactive reactions from the reference GEM, including the complete metabolic reaction set annotated by the genome, based on omics data to generate context-specific models, such as tissue or organ-specific models. The second one is to use omics data as constraints to improve metabolic flux prediction. Subsequently, researchers introduce current methods of using GEMs to analyze single-cell data, which are similar to those used for analyzing bulk data.
"We summarize the challenges, mainly stemming from the limitations of single-cell technologies themselves," said Dr. CHEN, the first author of this paper.
While GEMs are mainly used to analyze single-cell RNAseq data, with the development of single-cell proteomics and metabolomics, it is believed that GEM-aided analysis of multi-omics of single cells will come true in the near future.
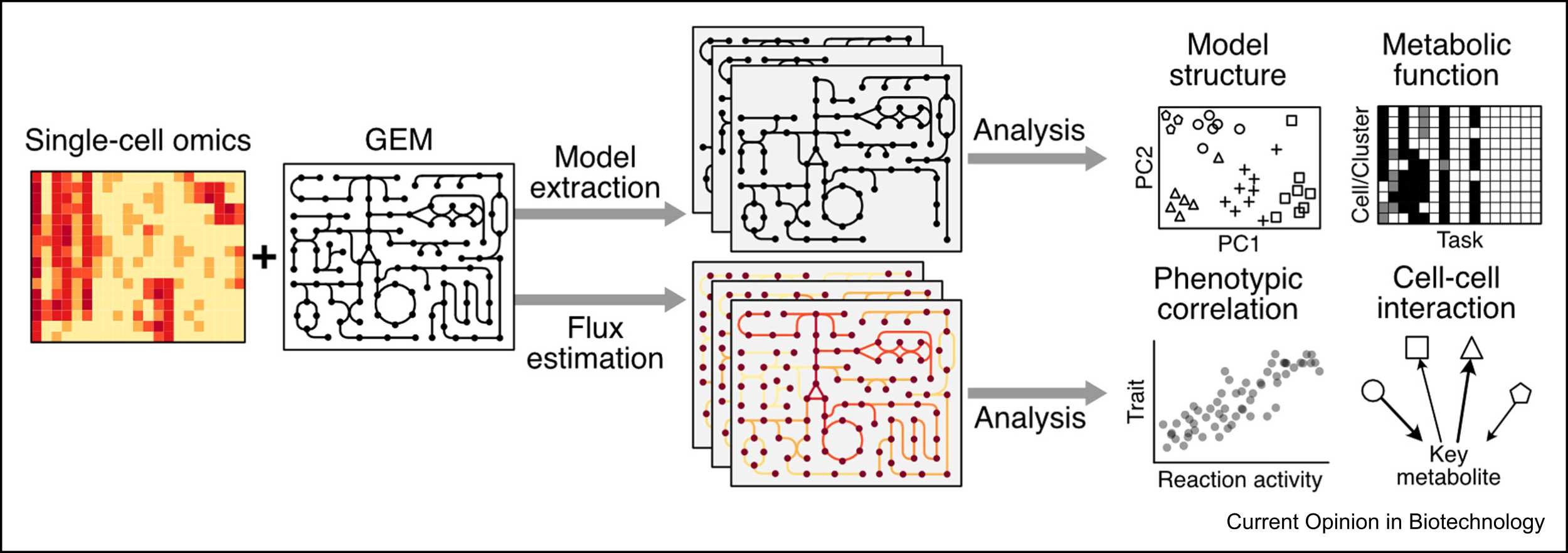
Single-cell omics analysis with GEMs. (Image by SIAT)
Media Contact:
ZHANG Xiaomin
Email:xm.zhang@siat.ac.cn