Researchers Reveal New Role of Cohesin Complex in Controlling Gene Coexpression
Date:29-08-2024 | 【Print】 【close】
In a study published in Nature Genetics on July 24, scientists from the Shenzhen Institute of Advanced Technology Chinese Academy of Sciences and the Howard Hughes Medical Institute/Janelia Research Campus, have discovered an unknown role of cohesin complex in controlling gene expression, thereby resolving the long-standing puzzle regarding the general function of genome structure in gene regulation.
The mammalian genome folds into high-order structures within three-dimensional nucleus, significantly influencing the establishment of chromatin landscape.Despite the well-characterized role of cohesin in genome organization, a perplexing finding is the lack of substantial gene expression changes at the cell population level after its depletion, raising ongoing debates about the relationship between genome structure and its biological function.
In this study, researchers measured transcriptome and chromatin accessibility at the single-cell level before and after cohesin loss, and developed a new statistical model based on Spearman’s rank-order correlation to quantify gene coexpression coefficients per pair of accessible chromatin domain.They found that instead of dictating population-wide gene expression levels, cohesin loss induces widespread gene coactivation and long-range chromatin co-opening incis, suggesting a default role to neutralize stochastic coexpression tendencies ofcis-linked genes in single cells.
To explore the mechanisms underlying this coregulation, the researchers performed genome-wide intron-sequential fluorescence in situ hybridization (FISH), lineage-specific single-molecule RNA FISH and live-cell protein imaging experiments. The results revealed that cohesin loss enhances gene co-bursting along the chromosome and blocks spatial mixing of transcriptional hubs.
Moreover, the analysis of live-cell single-molecules tracking further showed that cohesin limits the exploration of diverse enhancer- and core promoter-binding transcriptional regulators.
The researchers said these findings pinpointed an unknown layer of gene regulation – transcriptional co-activation. This new layer is achieved by gene grouping and sharing of transcriptional machinery resulting from genome folding and thus differs from the classical promoter-enhancer interaction mechanism.
“Our work could shed light on understanding the etiology of various cohesinopathy disorders, such as Cornelia de Lange syndrome, and might also provide insights into cell fate decision and developmental processes,” said DONG Peng, one of the corresponding authors of the study.
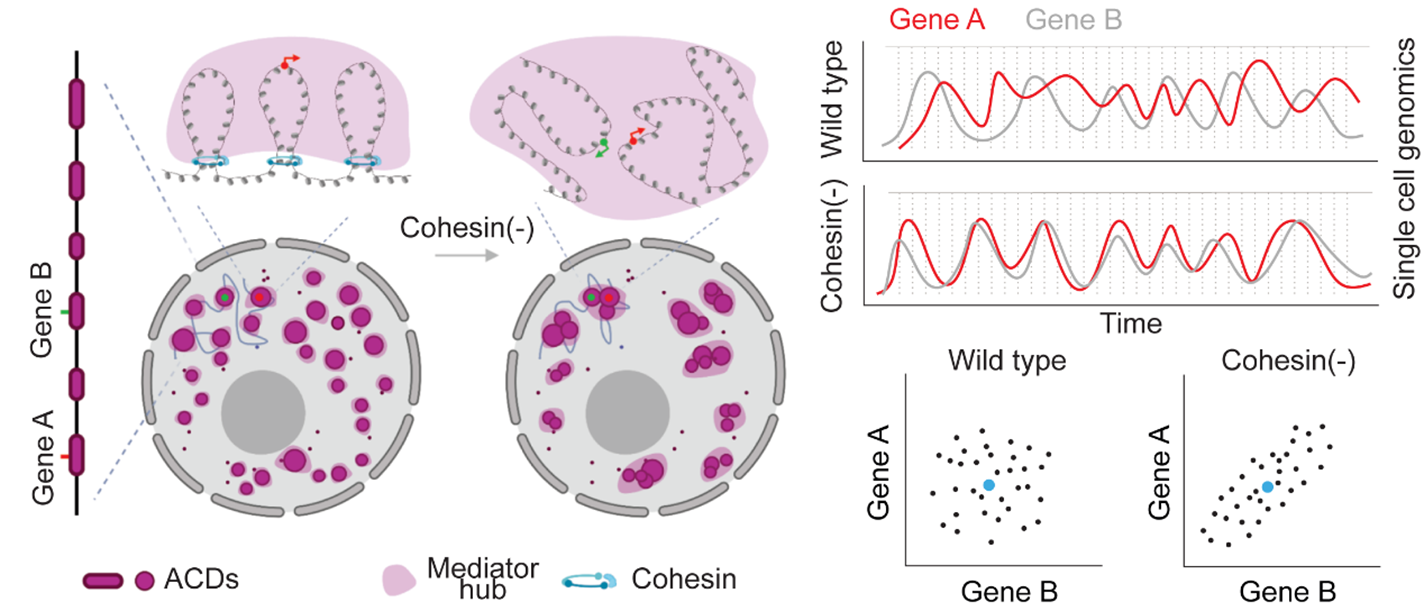
Transcriptional co-activation model controlled by genome folding (Image by DONG Peng)
Media Contact: LU Qun
Email: qun.lu@siat.ac.cn
Download the attachment: