
Novel Biological Signal-Processing Framework Precisely Decodes Complex Cellular Language
Cells naturally process external signals through intricate genetic circuits, enabling adaptation to diverse environments. These circuits rely on complex, nonlinear interactions among genes and proteins to regulate behavior, maintain homeostasis, and respond to environmental cues. Synthetic biology leverages these mechanisms to engineer biosensors, metabolic pathways, and therapeutic systems. However, engineering traditional synthetic circuits struggle to process complex biological signals effectively, as non-orthogonal signal responses hinder precise regulatory control.
Recently, a team led by Prof. CHEN Ye from the Shenzhen Institutes of Advanced Technology (SIAT) of the Chinese Academy of Sciences, in collaboration with Academician SHAN Yang of the Chinese Academy of Engineering, developed a scalable biological signal-processing framework that uses synthetic operational amplifiers" to convert mixed cellular inputs into clean, orthogonal outputs. This approach enables precise, predictable control of complex biological systems and paves the way for high-performance synthetic genetic regulation.
The study was published in Nature Communications on July 31.
Inspired by operational amplifiers in analog electronics, the researchers conceptualized cellular sensing as an "encoding" step: multiple signals generate a mixed, non-orthogonal transcriptional response. They then designed synthetic biological operational amplifiers (OAs) as "decoders", performing weighted subtraction and amplification to extract target components from composite signals.
The OA circuits rely on engineered transcription factors and promoters, combined with a mathematical model that treats signal transformation as a matrix operation. This design can be generalized from two-dimensional signal separation to N dimensions, offering scalability for highly complex biological networks.
The framework enabled the construction of multiple inducer-free, growth-stage-responsive circuits in Escherichia coli, achieving regulatory signal amplification of up to 153/688-fold, and was subsequently validated through two representative applications. In dynamic control of biomanufacturing, it enabled Escherichia coli to sense its growth phase and autonomously switch gene expression—promoting biomass accumulation during growth and producing target metabolites such as shikimic acid during production—without costly inducers, improving efficiency and reducing cost. In signal decomposition, it resolved complex crosstalk among three quorum-sensing signals, separating mixed inputs into three independent orthogonal outputs, showcasing strong performance in high-dimensional, complex signal processing.
This study establishes a foundational framework for synthetic biology by decoding cellular states, enabling precise control of biological systems. This advance offers solutions to key biomanufacturing challenges and paves the way for smarter, more robust cellular computers in medicine, environmental remediation, and sustainable energy.
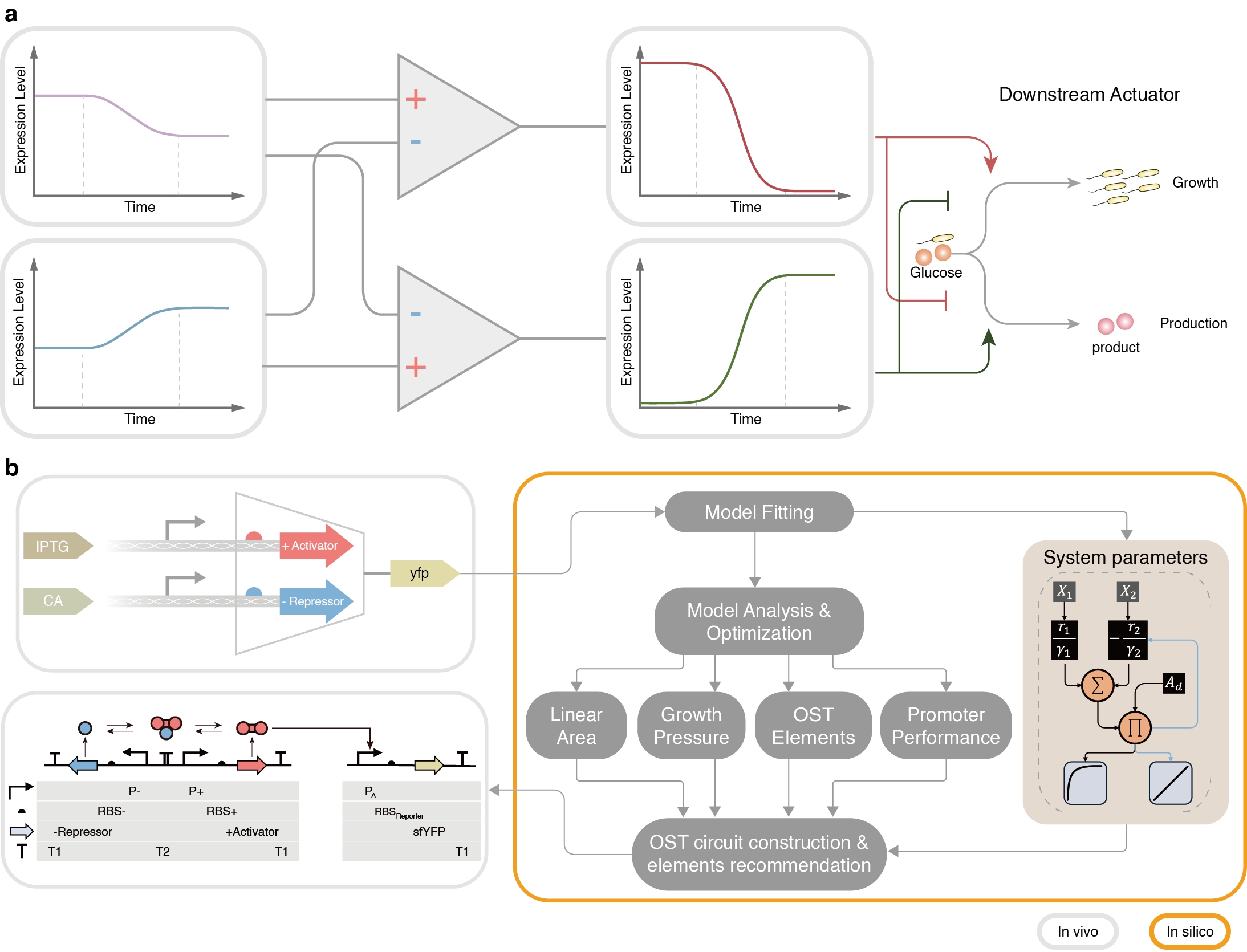
Schematic diagram of operational amplifier design and applications. (Image by SIAT)
File Download:
