
An AI-Driven Atomic Force Microscopy Platform for Decoding Human Immune Cell Mechanics
Macrophages drive key immune processes—including inflammation, tissue repair, and tumorigenesis—via distinct polarization states (e.g., M0, M1, M2), whose accurate identification is vital for diagnosis and immunotherapy. However, conventional methods like RNA sequencing and flow cytometry are often costly, time-consuming, and unable to enable real-time, label-free, high-throughput detection.
Atomic force microscopy (AFM) has emerged as a powerful tool in cell phenotyping by decoding the mechanobiological signatures of different cellular states, while AI enables rapid analysis of its complex data. Yet, macrophages remain underexplored in this context.
In a study published in Small Methods, a research team led by Prof. LI Yang from the Shenzhen Institutes of Advanced Technology (SIAT) of the Chinese Academy of Sciences, has developed and validated a label-free, non-invasive method combining AFM with deep learning for accurate profiling of human macrophage mechanophenotypes and rapid identification of their polarization states.
In this study, researchers used localized force-distance curves from AFM to extract biomechanical classifiers, then trained a deep neural network incorporating smart weight assignment and pixel voting to predict macrophage polarization states—naïve (M0), pro-inflammatory (M1), and anti-inflammatory (M2).
To validate the AI model, researchers analyzed the entire population of stimulated macrophages using a weighted voting algorithm, originally trained and optimized on well-characterized naïve M0, M1, and M2 phenotypes. The final probability distribution across the four categories—naïve M0, M1, M2, and M1/M2—was 4.3%, 52.2%, 26.1%, and 17.4% respectively.
Validation was subsequently performed using flow cytometry. The results indicate that pseudovirus stimulation predominantly induced an M1 phenotype (68.3%), with smaller proportions of M2 and mixed M1/M2 cells (15.5% and 15.1%, respectively), while naïve M0 cells were nearly absent (1.1%). The flow cytometry data largely corroborated the classifications generated by the AI model, underscoring its promising ability to distinguish macrophage subtypes, including mixed phenotypes.
This study offers a powerful tool for probing disease progression and therapeutic responses, which, as Prof. LI highlighted, can be extended beyond macrophages to other cell types. This capability paves the way for diagnostics based on mechanophenotypes in cancer, fibrosis, and infectious diseases.
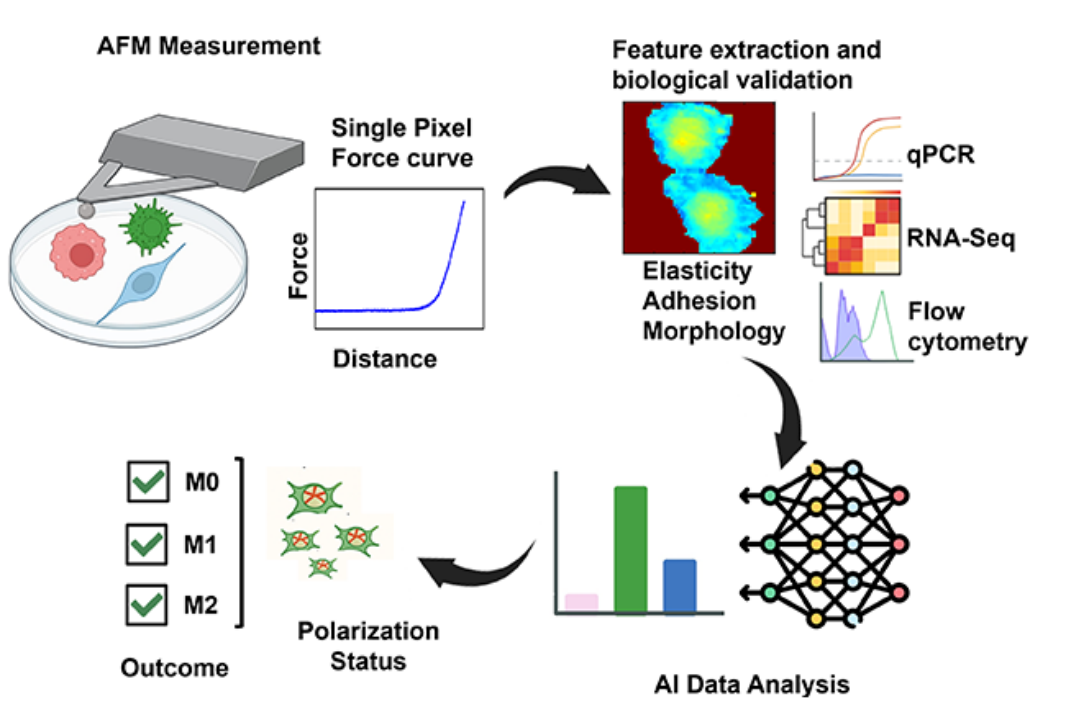
Using localized force-distance curves from AFM, a deep neural network was trained and biologically validated to predict and accurately distinguish macrophage polarization states, including complex states in macrophages. (Image by SIAT)
File Download:
